Week 5 Problem sets
PSET #5
ASSIGNED: Friday 08 May 2020
DUE: 5:00p Pacific 15 May 2020
NOTES:
- Given the unique circumstance of Spring 2020, we ask you to do your best to maximize your learning. Each problem set is an opportunity to assess your learning, identify gaps, reflect on what you have learned, and determine what you wish to learn next.
- You can discuss the PSET questions with other students. But your answers should be your own work. Please let us know if you collaborated with other students working on the PSET.
- Please turn in your completed problem sets as an electronic copy via Gradescope. Please make sure to clearly indicate the starting and ending boundaries of your answers to each question on Gradescope.
- Please do not go over any word limits.
- Please type your answers.
- Please label and provide descriptions for the figures.
- Please show your work and include units.
GOAL: Welcome to PSET-5. This problem set will help you describe how DNA read and write work. You will examine the consequences of non-trivial quantitative pacing in the development of these technologies. Finally, you will examine the consequence of interconvertability of matter and information as DNA read and write tools improve and discuss the qualitative changes that emerge.
(Q1) Sequencing the Total DNA in the Biosphere (20 pts)
Q.1.a The following paper estimates that there are about 5.3 * 10^37 bases of DNA in the biosphere.
Given the increasing trends in productivity of sequencing. In what century can a team of 10 sequence 5.3 * 10^37 bases in a month? We will use the information from Carlson Curves to provide a projection.
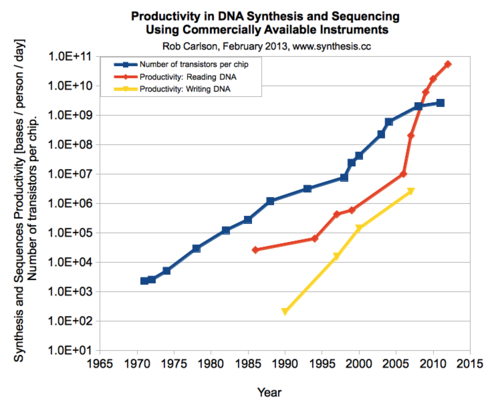
There are many ways to solve this problem. You can use your method or use the suggested assumptions and steps. (Your final answer depends on your assumptions).
Assumption.1 assumes the trend in sequencing productivity since 2005 will continue.
-
estimate 2005 productivity at 10^7 (bases per person per day)
-
estimate 2010 productivity at 10^10 (bases per person per day)
-
from above numbers produtivity rate (R) for every 5 years, 1000x better
Assumption.2 from the graph, we can estimate our current productivity P0 = 10^11 (bases per person per day) for the year 2015. (Note: The trend we are assuming (that every five years, the rate increases by 10^3) does not fit the actual data perfectly. That is ok for our estimate. Although 2015 should have a rate of 10^13 according to our assumption, we will use a P0 of 10^11 as shown on the graph.)
Step.1 (you have to do):
Normalize total DNA (5.3 * 10^37) bases to our team size (10) and time (30 days) to calculate expected productivity (P) with units (bases per person per day)
$P= \frac{total DNA}{team Size * 30 days}$
Step.2 (you have to do):
Construct a function that relates expected productivity to the productivity rate, current productivity, and time.
- Expected productivity: P (bases per person per day)
- Current productivity: P0 (bases per person per day)
- Time: t (number of five year periods)
- Current productivity rate: R (=1000, no units)
$ P = function (P0, R, t) $
$ P = P0 * R^t $
Then solve for t (number of five year periods)
Step.3 (you have to do):
Change t form number of five year periods to years. Add to initial time starting at (2015).
Q.1. Extra activity Construct a table comparing expected time for R: (1000, 2500, 5000, and 10,000). Construct a second table where t is number of two year periods instead of five.
(Q2) DNA as a Storage Medium (30 pts)
The US government has been using an underground salt mine in Pennsylvania as a long term storage facility for physical documents, primarily retirement-related papers. According to the Washington Post, in this facility, approximately 1000 employees pass thousands of case files from cavern to cavern and then type in retirees’ data, one line at a time. Basic document retrieval involves a forklift and takes days.
Given the trends in DNA read and write, consider using DNA as an alternative storage medium for these documents.
Helpful numbers:
- Volume of storage facility = 10 million m^3
- Letters per cubic meter of typical government document = 10^9 letters/m^3
- Volume of a DNA base pair ~ 10^-27 m^3
Q.2.a. Since DNA has only 4 bases, but English has 26 letters, you will somehow have to map letters to bases. Briefly specify an encoding scheme for mapping bases into the 26 English letters. (You do not have to give a letter to DNA mapping for all letters. The first few will do the trick.)
-
Hint-1: In your encoding scheme, the length of the bases should be consistent for the entire mapping.
-
Hint-2: What is the minimum number of bases you need to map the entire 26 English letters. Consider what would be the problem if you pick 2 bases (AA, TA, CA, GA,…) to map 26 letters.
Q.2.b. Using your letter to DNA mapping, how many total base pairs of DNA would be needed to store the archival contents of the salt mine. Assume that the salt mine is completely packed with documents.
Q.2.c. What would be the physical volume of DNA encoding the salt mine documents?
Will the DNA fit into a thimble? Yes / No
Q.2.d. How expensive would it be to print the DNA (assume cost ~1 cents/base)? Compare your number against the United States GDP (20.54 trillion USD - 2018)
Q.2.e. Give two advantages and two disadvantages of using DNA as a storage medium (4 bullet points)
Q.2.f. Can information be stored in the DNA in a living organism as opposed to DNA stored in a frozen tube (-80 freezer)? If yes, what are one advantage and one disadvantage of using live-cell storage? (2 bullet points)
Helpful resources:
(Q3) Cost of Education vs Cost of Germline Editing (20 pts)
Over the past 12 years, the price of synthesizing genes has dropped from 4 USD to 0.04 USD per base pair. Presume the future price of DNA synthesis will continue to drop two-fold every two years.
Stanford’s undergraduate tuition is approximately 50,000 USD per year, up from 25,000 USD in 2000.
Presume Stanford’s tuition will continue to double every 15 years.
Q.3.a If a human genome is 4 billion base pairs long, then when will the cost of synthesizing the DNA encoding an entire human genome be roughly the same as the tuition cost associated with attending Stanford for one year?
Hint: Use the facts given. Keep your math simple. Show your work. An approximate answer is fine.
Q.3.b What would be your estimate if the price of synthesizing genes continue to drop two-fold every 2.5 years instead of 2 years (i.e., only a 6-month longer doubling time)? An approximate answer is fine.
Q.3.c Given your answer from part (a) presume that you could then design, build, and bring to life a human encoded by a genome that you design (e.g., with any and all mutations that you and your partner choose). As potential parents, would you prefer to spend your savings on college tuition for a natural child or on realizing a genetically engineered offspring who then has to make their own way? Why? (2-3 sentences)
(Q4) Paper reading activity (30 pts)
First examine, Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform or here
Q.4.a In your own words, what is the primary claim of the paper? What are the primary evidence in support of the claim? (2-3 sentences)
Q.4.b. What is your reaction and thoughts to this work? What are the benefits and risks of this work? Do the benefits of the paper outweigh its risks? Should this work be published? Think back to Rousseau and Hobbes’s discussion from Week-2, and consider the interconvertability of DNA as matter and information. (3-4 sentences)
(We wish to hear your thoughts, and we don’t have a “correct answer” in mind. Instead, we want you to think deeply and practice your written communication skills.)
Next examine,Design and synthesis of a minimal bacterial genome.
Q.4.c In your own words, what is the primary claim of the paper? What are the primary evidence in support of the claim? (2-3 sentences)
Q.4.d What were the genome size and number of genes in JCV-syn3.0? How many were essential genes of previously unknown function discovered in this work? How is the whole genome synthesized in this work?
Final Project - Group Activity.1
Brainstorm report + Team Rules
ASSIGNED: Friday 08 May 2020
DUE: 5:00p Pacific 15 May 2020
NOTES:
- Include your group number and names of your group members.
- Team Rules is 6 points of your Final Project.
- Brainstorm is 5 points of your Final Project.
- Please turn in a single (one person per team) electronic copy of your Group Activity.1 via Gradescope.
Q.1. What is your group number? (If you have a group name - let us know)
Q.2. Turn in a copy of your team rules. We expect to see between 8 and 20 rules (8 rules < # rules < 20 rules) that you have all agreed upon. (6 points)
Q.3. Brainstorm report: As a group for your project brainstorm ideas. Brainstorm should include following three themes:
-
People health
-
Planet health
-
Political health
we expect 30 ideas for each theme total 90 ideas. If you have more ideas, please share them.
Extra learning (100% optional):
(Q5)Sequencing and Synthesis technologies (0 pts)
In the pre-class material, you were introduced to a DNA read method commonly referred to as next-generation sequencing (NGS) or massively parallel sequencing. Let’s examine an alternative approach, namely nanopore sequencing.
Watch this video
5.a. Describe how Nanopore sequencing works (use bullet points)
MinION is a product that uses Nanopores for sequencing. While the error rate with Nanopore technology is between 5-10%. additional Link. This tool can be used to sequence and assemble a human genome additional link. Now briefly examine the various product sequencing technology from Illumina link.
5.b. How are NGS and Nanopore sequencing different?
5.c. Imagine that the storage of information in DNA from the previous question has become a routine platform. You have been hired as a consultant for a library that seeks to store its archive inside the DNA medium. Your task is to determine suitable reading platforms. What type of“read technology” would you recommend: a sequencer that gives massive simultaneous short reads with low error, or fast readers that read long sequences with error rates as high as 15 %. Please include quantitative reasoning in your answer.
5.d. Where would you use an Illumina sequencing machine instead of a minION sequencing machine? And vice versa?
github source code for teaching staff