Week 8 Preclass
Evolution as Tool for Engineering
Please read and consider the below before start of class. The questions given are only study questions not homework to be graded. Talk about it all with your classmates, friends, or TAs, as you like.
No class on Monday
Preclass for Wednesday
From Natural Algorithm to Engineering Service
This week you will examine the role of evolution from a Bioengineering perspective. After considering the following example and the material throughout this week you will be able to:
Goal-1: Describe evolution as an algorithm that follows a set of rules.
Goal-2: Utilize evolution as an algorithm. “Evolution, at your service!”:
This robust algorithm is a service that can be deployed in diverse contexts.
Goal-3: Describe the limitations when used as a service, and begin to describe what evolution could look like in the future (“Evolution 2.0”).
The Core concept for this week includes: Evolution: Population, Diversity, Fitness, Selection (Pressure), Mechanics (Random Mutations), and Algorithm vs Service.
Before we begin briefly explore these: link and link-2 to better appreciate these examples.
Example-1: Why do you need to get a flu shot every year?
To begin, let us consider 2018 flu season, which was considered as the worst flu season since 2009-2010.
Following is from an article published in January 2018:
“Every winter brings cautionary tales that the flu—just the regular old flu—can kill. And the cautionary tales this year are hard to beat. Twenty-one-year-old Kyler Baughman, for example, a fitness buff who liked to show off his six-pack, recently died a few days after getting a runny nose.” source
According to the numbers, this (2018) year’s flu season is in fact worse than usual. It got started early, and it’s been more severe. Twenty kids have died of the flu since October. And in the week ending January 6, 22.7 out of every 100,000 hospitalizations in the U.S. were for flu—twice the number of the previous week.”
Every year, we are reminded to go get a flu shot.
Q.1. If you got a flu vaccine last year, why do you still have to get a flu shot again this year?
Q.2. What are the two populations that are interacting in the above story and graph?
Q.3. How do new flu strains arise every year? Where do they come from?
Q.4. What causes the flu virus to change?
Briefly examine the two following links:
Let’s highlight to the following from above link: “Tiny mutations help the virus evade detection, and as it evades detection, it’s more successful at multiplying.”
An algorithm can be defined as:
“A process or set of rules to be followed in calculations or other problem-solving operations, especially by a computer.” (via Google)
Given this definition, think about evolution in this new context.
Q.5. If evolution is an algorithm, what problem is being solved, and by whom (or what)? What are the rules?
Extracting out the idea of Evolution from this example evolution is the results of two intertwined processes:
-
Random Mutations beget genetic variation; non-random selection to improve “fitness”.
-
The end result may look non-random, but the mechanism that enables evolution and natural selection to occur is entirely random.
The example of flu virus highlights the algorithm of evolution acting on a timescale that is easy for us to understand.
Example-2: Ancient virus genomes reveal complex evolution of Hepatitis B
In a fascinating research article see also (article), “Scientists reported on Wednesday that they have recovered DNA from the oldest viruses known to have infected humans — and have succeeded in resurrecting some of them in the laboratory. The viruses were all strains of hepatitis B. Two teams of researchers independently discovered its DNA in 15 ancient skeletons, the oldest a farmer who lived 7,000 years ago in what is now Germany. Until now, the oldest viral DNA ever recovered from human remains was just 450 years old.”
Here is the abstract from the article:
“The hepatitis B virus (HBV) is one of the most widespread human pathogens known today, yet its origin and evolutionary history are still unclear and controversial. Here, we report the analysis of three ancient HBV genomes recovered from human skeletons found at three different archaeological sites in Germany.
We reconstructed two Neolithic and one medieval HBV genomes by de novo assembly from shotgun DNA sequencing data. Additionally, we observed HBV-specific peptides using paleo-proteomics. Our results show that HBV circulates in the European population for at least 7000 years. The Neolithic HBV genomes show a high genomic similarity to each other. In a phylogenetic network, they do not group with any human-associated HBV genome and are most closely related to those infecting African non-human primates. These ancient virus forms appear to represent distinct lineages that have no close relatives today and went possibly extinct.
Our results reveal the great potential of ancient DNA from human skeletons in order to study the long-time evolution of blood borne viruses.”
Note the highlighted lines from the abstract.
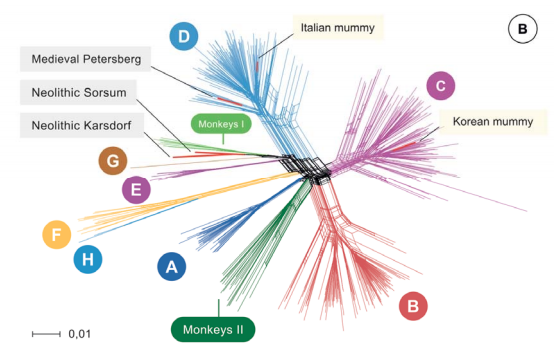
Figure-1 Network of 495 modern, two published ancient genomes (light yellow box), and three ancient hepatitis B virus (HBV) obtained in this study (grey box). Colors indicating the 8 human HBV Genotypes (A-H), two monkey genotypes (Monkeys I, African apes and Monkeys II, Asian monkeys) and ancient genomes (red).research article
Q.6. What is the role of evolution (as an algorithm) in this example?
Summary
Through the two examples, you have started to synthesize the workings of evolution and natural selection as an algorithm. This week you will engage with this algorithm as a tool (and service) that can be used to solve challenges in science and engineering. In doing so, you will start to identify the current limitations of evolution, and consider what it would mean to “improve” evolution.
Additional Resources: Please make sure to briefly take a look at these great resources:
1. Collection of links - excellent place to learn about fundamental concepts in evolution
2. Misconceptions about evolution
3. Host and Viral Evolution: Molecular Evolutionary Arms Race Between Primate and Viral Genomes link
4. Spanish Influenza Pandemic and Vaccines
Preclass for Friday
In the previous class you were introduced to evolution as an algorithm. Next, you will examine the role of evolution as an engineering solution and approach. After considering the following example and the material throughout this week you will be able to:
Goal-1: Abstract evolution from a population-wide biological force to a service that can be deployed by a group or an individual. (“Evolution, at your service!”)
Goal-2: Be able to identify when evolution is a good engineering choice and why? (also begin to address who gets to decide?)
Goal-3: Understand the limitations and benefits of this approach.
Example-1: “Evolution, at your service!”
Since the days of Gregor Mendel, geneticists (and avid gardeners/monks) have understood that we can breed populations for certain traits. By taking advantage of our understanding of heritability and genetics, we can - for example - make chickens with extravagant traits see here: (link, link)
Another example of human intervention in evolution is dog breeding. Take a look at the link to see the evolution of Petface here From the article: Cranial length, snout length and skull length all contribute to the difficulties brachy dogs have in breathing. (PLOS One)
Q.1. Think back to the pre-class material for Wednesday. What are the selection pressures (and their sources) in the case of flu and dog/chicken breeding? How is the fitness determined in each case?
Q.2. Thinking of evolution as an algorithm, where else can we intervene?
Example-2: Directed evolution of proteins
Directed evolution is an effective strategy for improving or altering the activity of biomolecules for industrial, research and therapeutic applications.
Laboratory evolution of proteins requires methods for generating genetic diversity coupled with identifying protein variants with desired properties. (You can read on the scientific background on the Nobel Prize in Chemistry 2018 awarded to the Directed Evolution of enzymes and binding proteins from Link)
Here is an introduction from Methods for the directed evolution of proteins
“ Over many generations, iterated mutation and natural selection during biological evolution provide solutions for challenges that organisms face in the natural world. However, the traits that result from natural selection only occasionally overlap with features of organisms and biomolecules that are sought by humans. To guide evolution to access useful phenotypes more frequently, humans for centuries have used artificial selection, beginning with the selective breeding of crops and domestication of animals.
More recently, directed evolution in the laboratory has proved to be a highly effective and broadly applicable framework for optimizing or altering the activities of individual genes and gene products, which are the fundamental units of biology Genetic diversity fuels both natural and laboratory evolution. The occurrence rate of spontaneous mutations is generally insufficient to access desired gene variants on a time scale that is practical for laboratory evolution. A number of genetic diversification techniques are therefore used to generate libraries of gene variants that accelerate the exploration of a gene’s sequence space.
Methods to identify and isolate library members with desired properties are a second crucial component of laboratory evolution. During organismal evolution, phenotype and genotype are intrinsically coupled within each organism. However, during laboratory evolution, it is often inconvenient or impossible to manipulate genes and gene products in a coupled manner. Therefore, single-gene evolution in the laboratory requires carefully designed strategies for screening or selecting functional variants in ways that maintain the genotype–phenotype association.”
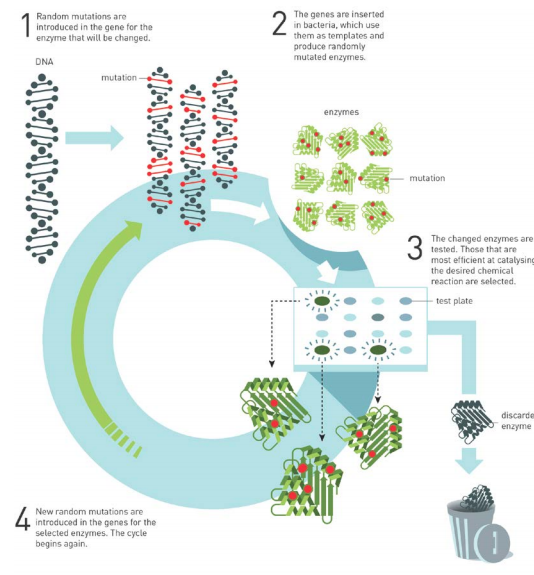
Figure-1 The work flow for the directed evolution of enzymes. Source
Natural selection: A process by which individuals with the highest reproductive fitness pass on their genetic material to their offspring, thus maintaining and enriching heritable traits that are adaptive to the natural environment.
Artificial selection: A process by which human intervention in the reproductive cycle imposes a selection pressure for phenotypic traits desired by the breeder. (Also known as selective breeding).
Libraries: Diverse populations of DNA fragments that are subject to downstream screening and selection.
Q.3. How can directed evolution be used an approach to evolve desired proteins?
Q.4. What are the challenges associated with this approach?
Q.5. When is directed evolution a good choice?
Example-3: Antibiotic resistance
Antibiotic resistance and the threat of drug resistance bacteria is an emerging challenge that grows every year. (link-1, link-2).
According to CDC:
“Overuse and misuse of antibiotics can promote the development of antibiotic resistance. Every time a person takes antibiotics, sensitive bacteria (bacteria that antibiotics can still attack) are killed, but resistant bacteria are left to grow and multiply.
This is how repeated use of antibiotics can increase the number of drug-resistant bacteria. Antibiotics are not effective against viral infections like the common cold, flu, most sore throats, bronchitis, and many sinus and ear infections. Widespread use of antibiotics for these illnesses is an example of how overuse of antibiotics can promote the spread of antibiotic resistance. Smart use of antibiotics is key to controlling the spread of resistance.”
Q.6. What is the role of evolution in antibiotic resistance?
To answer the question above recall that in an earlier module you came across the MEGA-plate. The (120 × 60 centimeters) plate on which bacteria spread and evolved on a large antibiotic landscape (Mega-plate video)
Here is the abstract from the paper:
“A key aspect of bacterial survival is the ability to evolve while migrating across spatially varying environmental challenges. Laboratory experiments, however, often study evolution in well-mixed systems. Here, we introduce an experimental device, the microbial evolution and growth arena (MEGA)–plate, in which bacteria spread and evolved on a large antibiotic landscape (120 × 60 centimeters) that allowed visual observation of mutation and selection in a migrating bacterial front. While resistance increased consistently, multiple coexisting lineages diversified both phenotypically and genotypically. Analyzing mutants at and behind the propagating front, we found that evolution is not always led by the most resistant mutants; highly resistant mutants may be trapped behind more sensitive lineages. The MEGA-plate provides a versatile platform for studying microbial adaption and directly visualizing evolutionary dynamics.”
Q.7. Could we use evolution to combat antibiotic resistance?
Useful Additional Resource: Review the key Takeaways from: Methods for the directed evolution of proteins
github source code for teaching staff